The CAMDA Challenges
As traditional in CAMDA contests, neither we nor the producers of the data can provide advice on the datasets to individuals as dealing with the files forms part of the analysis challenge. There is, however, an open forum for the free discussion of the contest data sets and their analysis, in which you are encouraged to participate. For CAMDA 2015, we have have compiled the following exciting contests:
- The FDA SEQC consortium has compiled a series of synthetic benchmarks and applied use-cases to assess the performance of modern gene transcript expression profiling methods, for the first time systematically assessing RNA-Seq in a wider context. SEQC is providing two benchmark challenges to CAMDA 2015:
- A toxicogenomics study with matched NGS and microarray profiles for the response of over 100 rat livers to 27 chemicals with 9 different modes of action.
- A synthetic reference benchmark with built-in truths, spanning multiple NGS, microarray, and qPCR platforms.
- A selection of large-scale cancer studies of less well-studied diseases from the lastest data release of the International Cancer Genome Consortium (ICGC), including matched gene and microRNA expression profiles from RNA-Seq, somatic CNV, methylation, and protein expression profiles. Only processed data are provided due to access restrictions by ICGC.
Please notice that CAMDA challenges are not limited to questions proposed here. We look forward to a lively contest!
FDA SEQC Challenges
The FDA SEQC consortium has compiled a series of synthetic benchmarks and applied use-cases to assess the performance of modern gene transcript expression profiling methods, for the first time systematically assessing RNA-Seq in a wider context. SEQC is providing two benchmark challenges to CAMDA 2015.
References:
- The MAQC/SEQC Consortium - Z. Su, PP. Łabaj, S. Li, (2014). A comprehensive assessment of RNA-seq accuracy, reproducibility and information content by the Sequence Quality Control consortium. Nat. Biotechnol. 32, 903-914.(link to article)
- S. Li et al., C. (2014). Detecting and Ameliorating Systematic Variation from Large-scale RNA Sequencing. Nat. Biotechnol. 32, 888-895. (link to article)
- C. Wang et al. (2014). A comprehensive study design reveals treatment- and abundance-dependent concordance between RNA-Seq and microarray data., Nat. Biotechnol. 32, 926-932. (link to article)
Challenge 1a: SEQC Rat TGx - rat liver response to chemicals
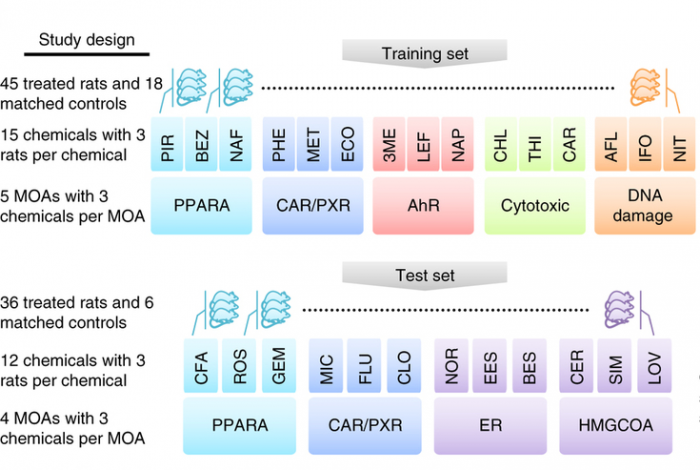
In this study, matched RNA-Seq and microarray gene expression profiles were collected of 105 rat livers to test their response to 27 chemicals representing 9 different modes of action (MOA). The NGS reads collected comprise 1.5 Terabases. In the study, a key question was the predictability of the chemical mode of action. Initial platform comparison showed consensus as well as variation, and effects of data processing were not yet further explored.
Data Description
This data comprised a training set and a test set with the text on the left detailing the experimental design and the text on the right listing the key analyses conducted (see figure below). Both microarray and RNA-seq were used to profile transcriptional responses induced by treatment of rats by each chemical; each is associated with a specific mode of action (MOA). For each MOA there were three representative chemicals and three biological replicates per chemical. Cross-platform concordance was evaluated at multiple levels: deferentially expressed genes, mechanistic pathways and MOAs. To compare the predictive potential of RNA-seq and microarray as gene-expression biomarkers, four MOAs by both platforms were analyzed as a test set. Two of the MOAs (PPARA and CAR/PXR) were present in the training set whereas the other two were not.
Questions of interest include, but are not limited to
- Topic 1: Inter-platform concordance: Is a shared list of genes the ‘golden standard’ to test the cross-platform consistency of the underlying biology? What biological insights are unique to data from one of the two platforms? Can we understand or adjust for that?
- Topic 2: Classification / prediction: We know we can get 100% accurate prediction of the chemical mode of action for RNA-Seq. Can you develop a similarly good predictor that works cross-platform?
- Topic 3: Alternative splicing: What can we learn about the role of alternative splicing from the RNA-Seq data?
Data download For this challenge, raw and processed data are provided as separate packages. The data packages contain metadata files, and either processed or raw data folders. Participants who want to use this dataset should read and accept the data download agreement to get access.
Challenge 1b: SEQC Main Study - Synthetic reference benchmark
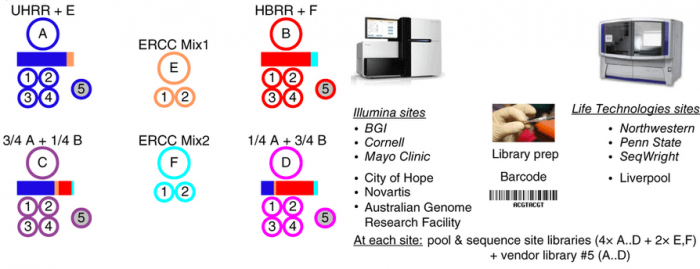
In this study, we present a reference benchmark data set comprising 30 billion reads (3 Terabases) of RNA-Seq data from The Sequencing Quality Control (SEQC) project, coordinated by the US Food and Drug Administration. Centrally prepared mRNA materials mixed from reference RNA samples with built-in controls were distributed to several independent sites for the construction of four replicate RNA-seq libraries per sample. Those four replicates, plus a fifth vendor prepared library distributed to the three ‘official sites’ for each platform, were then sequences. Platforms include Illumina’s HiSeq 2000 and Life Technologies’ SOLiD 5500 instruments.
Data Description
This dataset contains on a group of studies assessing different sequencing platforms in real-world use cases, including transcriptome annotation and other research applications, as well as clinical settings. In the main study RNA samples A to D were analyzed. Samples C and D were created by mixing the well-characterized samples A and B in 3:1 and 1:3 ratios, respectively. This allows tests for titration consistency and the correct recovery of the known mixing ratios. Synthetic RNAs from ERCC were both added to samples A and B before mixing and also sequenced separately to assess dynamic range (samples E and F). Samples were distributed to independent sites for RNA-seq library construction and profiling by Illumina's HiSeq 2000 (three official + three unofficial sites) and Life Technologies' SOLiD 5500 (three official sites + one unofficial site). Unless mentioned otherwise, data show results from the three official sites (italics). In addition to the four replicate libraries each for samples A to D per site, for each platform, one vendor-prepared library A5…D5 was being sequenced at the official sites, giving a total of 120 libraries. At each site, every library has a unique bar-code sequence, and all libraries were pooled before sequencing, so each lane was sequencing the same material, allowing a study of lane-specific effects. To support a later assessment of gene models, we sequenced samples A and B by Roche 454 (3×, no replicates, see Supplementary Notes to ref. 1, section 2.5).
Questions of interest include, but are not limited to
- Topic 1: Effect size versus sensitivity: How does the sample mixing ratio affect our ability to identify tissue specific signatures and markers?
- Topic 2: Signal-level models: Can we improve models of noise and bias for better site to site reproducibility?
- Topic 3: Junction / alternative transcript discovery: (i) Looking across sites and platforms, can we improve on the state of the art for junction / alternative transcript discovery (incl. consistency of novel junctions reads and coverage of the flanking exons)? (ii) Can we leverage the ultra-deep sequencing data to characterize performance as a function of read depth? (iii) Characterization of pipeline specific novel junctions
- Topic 4: What metrics can we develop by use of ‘built-in truths’ that are more sensitive to (i) a background signal, e.g., contamination by genomic DNA or a high prior belief of expression? (ii) consistent bias/error, e.g., a read aligner always just picking the first perfect match along a genome?
Data download For this challenge, raw and processed data are provided as separate packages. The data packages contain metadata files, and either processed or raw data folders. Participants who want to use this dataset should read and accept the data download agreement to get access.
ICGC Cancer Genome Consortium Challenges
From the comprehensive description of genomic, transcriptomic and epigenomic changes provided by ICGC, the main goal of this challenge is to gain novel biological insights to less well studied cancers selected here. However, we are not merely looking for 'old paradigm' cancer subtype classification!
Data Description and Download
For this challenge, only processed data are provided. These cancers all have matched gene expression, microRNA expression, protein expression profiles, somatic CNV, and methylation.
The above links point to release 17 and were meant as the official data set for the contest. We have received reports of the ICGC servers failing to provide particular files without corruption, however. Having no control over the ICGC servers we have, however, been able to download the corresponding files from release 18 instead. Base links for release 18:
Challenges
- Question 1: What are disease causal changes? Can the integration of comprehensive multi-track -omics data give a clear answer?
- Question 2: Can personalized medicine and rational drug treatment plans be derived from the data? And how can we validate them down the road?



















STAY CONNECTED
Tweet